EOS Tuning
First time reading about EOS tuning?
Check out our article on the topic here!
The goal of the EOS Tuning feature is to fine-tune the initial C7+ characterized EOS model to match PVT experiment data. This data consists of the standard PVT experiments like the constant composition expansion (CCE), and more complex experiments like the slimtube experiment. The EOS Tuning feature does not consider the viscosity experiments, as this is covered in the Viscosity Modeling feature.
The EOS Tuning feature has four main inputs that you can adjust to impact your predictions. The inputs are:
-
C7+ characterization case.
-
EOS parameter tuning.
-
Compositional adjustments
-
Weight factors.
All four of the inputs are covered in the sections below.
C7+ Characterization Case
You should always try to compare the different C7+ characterization cases you have saved! Unless you have distillation data with measured cut molecular weights, it can be hard to distinguish between the Twu and Effective Paraffin molecular weigtht models. The EOS tuning might shed some light on which model is most representative!
EOS Parameter Tuning
What would we recommend if this is your first time?
By default, only the BIPs are unlocked as regression in the tuning! If you don't get good results when running the EOS tuning case with just BIPs we recommend the following order of applying more tuning parameters:
-
Critical temperature (Tc) ramping.
-
Critical pressure (pc) ramping.
-
Critical temperature heaviest fraction (Tc) CN+ multiplier.
-
Critical pressure heaviest fraction (pc) CN+ multiplier.
-
Normal boiling point temperature (Tb) ramping.
-
Normal boiling point temperature heaviest fraction (Tb) CN+ multiplier.
EOS parameter tuning is a required step to develop an accurate model for petroleum mixtures with some or a lot of C7+ components because there are no truly predictive models that can estimate these parameters. The EOS parameters for the C7+ components that can be tuned in whitsonPVT are the following:
-
Critical temperature (Tc)
-
Critical pressure (pc)
-
Normal boiling point temperature (Tb)
-
Binary interaction parameters (kij) also called BIPs
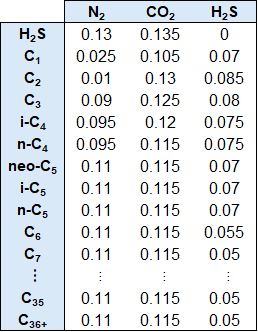
The reason why the acentric factors are not on the list above is because they can be uniquely determined from the critical point and the normal boiling point which are both more well constrained physical properties. The BIPs are scaled up / down using the Chueh-Prausnitz correlation between C1 and the C7+ SCN compoonents. The default model parameters for the Chueh-Prausnitz correlation are set to A=1 and B=1 for the Peng-Robinson 1979 (PR79) model and A=0 and B=1 for the Soave-Redlich-Kwong (SRK) model. For the non-hydrocarbon and hydrocarbon BIPs, a set of default values are used as shown in the table below.
Table 1: BIPs for non-hydrocarbon and hydrocarbon pairs.
There are some additional optionalities available in whitsonPVT, like choosing a different first component to start the tuning from (the default value is C7). You can also adjust the bounds by clicking the slider icon.
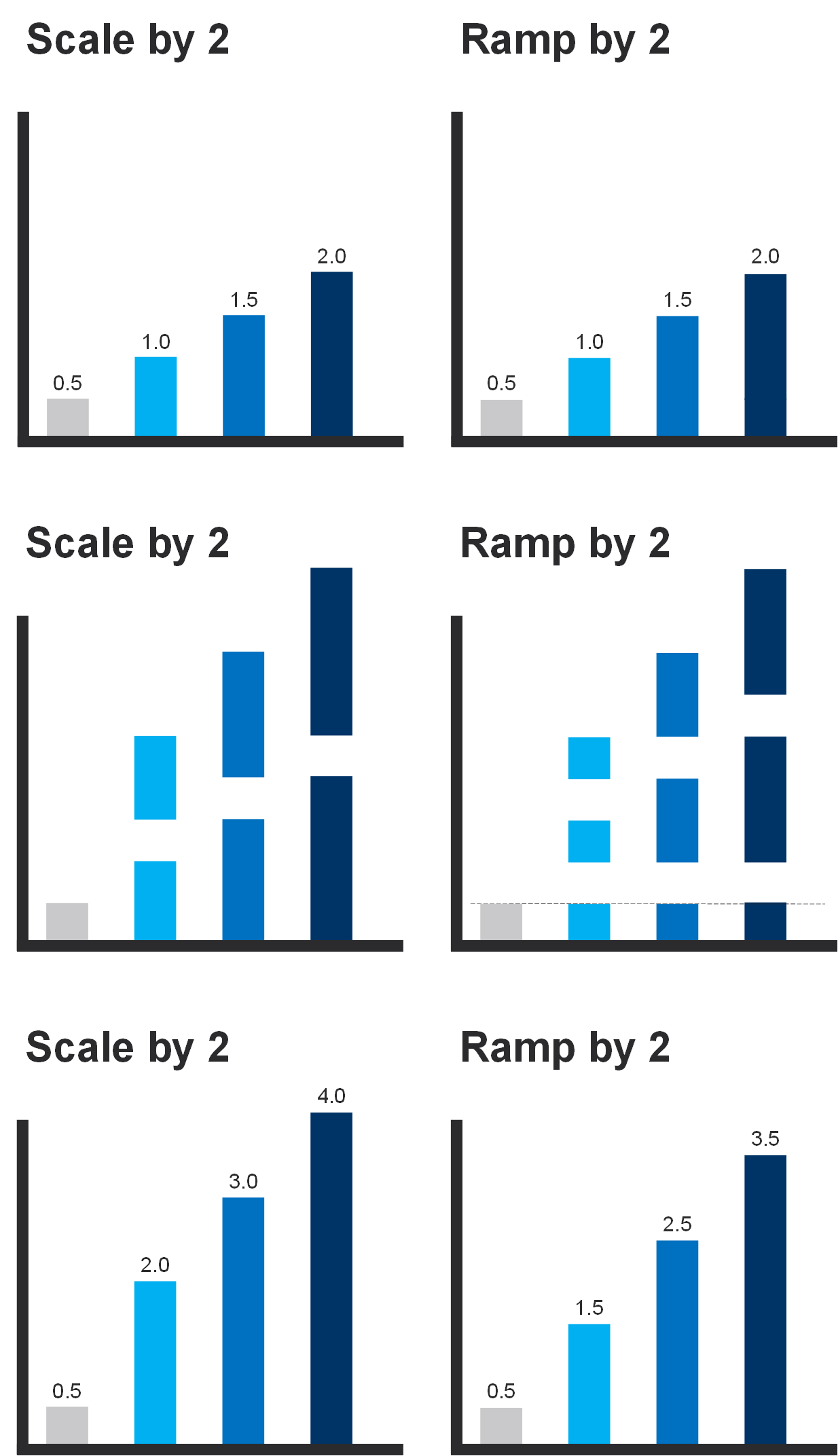
In whitsonPVT there are two methods for adjusting the C7+ EOS parameter values and an additional adjustment that can be applied to the heaviest fraction (i.e. the C36+ component). The two methods are ramping and scaling and are explained in detail below.
Ramping is a single regression parameter tuning approach that aims to honor the growing uncertainty in the component properties for the heavier components. The method can be defined in Equation \eqref{eq:ramping_definition}.
where \(\theta\) is the EOS property, the subscript \(n\) indicates the first plus component (usually set to 7), the subscript \(x\) indicates the C7+ component SCN component name (i.e. \(x \geq n\)).
Scaling is a single regression parameter tuning approach that simply sclaes all component properties up / down by a single scaling factor (\(S\)). The method can be defined in Equation \eqref{eq:scaling_definition}.
where \(\theta\) is the EOS property, the subscript \(x\) indicates the C7+ component SCN component name (i.e. \(x \geq n\)).
A comparison between ramping and scaling with a value of 2 for \(R\) and \(S\) is given in the figure below
Compositional Adjustments
What would we recommend if this is your first time?
Before trying to apply the compositional adjustments, try some different cases without compositional regression! If you are applying the compositional adjustment, check if most or all the data goes in the same direction of better predictions.
Some datasets that we recomment checking when applying compositional adjustments are:
-
Saturation pressure.
-
CCE single-phase density.
-
MSS density, formation volume factor, and gas removed.
-
DLE density, oil formation volume factor, and gas removed.
-
Iduvidual experiment CVD liquid dropout.
Compositional errors are real and can have a significant impact on your phase behavior predictions! However, with great power comes great resposibility. What do we mean by this? It is easy to match certain key predictions like saturation pressure, with very slight compositional adjustments, but this is not always the correct option! In general, we recommend trying changing the C7+ charaterization case (e.g. using Twu or Effective Paraffin MWs) and EOS tuning first before you adjust the composition, if you are going to change the composition
The two most common types of compositional errors that should be adjusted are:
-
Recombination GOR.
-
Average molecular weight.
Both of these options are possible to adjust in whitsonPVT! The recombination GOR adjustment lets you adjust the proportion of the flashed gas and oil in the recombined fluid. The default constraint in whitsonPVT is to constrain the compositional adjustment in the C1 mole content by 1 mole%. Round-robin studies have shown recombinations varying up to 3 mole%. The average molecular weight lets you adjust the molecular weight used in the gamma model and can be either the flashed oil or recombined fluid molecular weight. The choice of fluid used is based on the availablity of compositional data. The priority goes (1) flashed oil, (2) separator oil, (3) recombined fluid.
The compositional adjustment is done induvidually for each sample in the EOS tuning.
Weight Factor Cases
What would we recommend if this is your first time?
This is an advanced feature, and unless you know what data you want to priritize, we recommend that you stick to the default case!
Weight factor cases can be added to emphasize certain properties in the different PVT experiments. When making a custom weight factor case, you can choose a basis which automatically fills the property weights with the basis case's values. This lets you make slight adjustments without needing to re-enter all the data.
EOS Tuning Results and Predictions
What would we recommend if this is your first time?
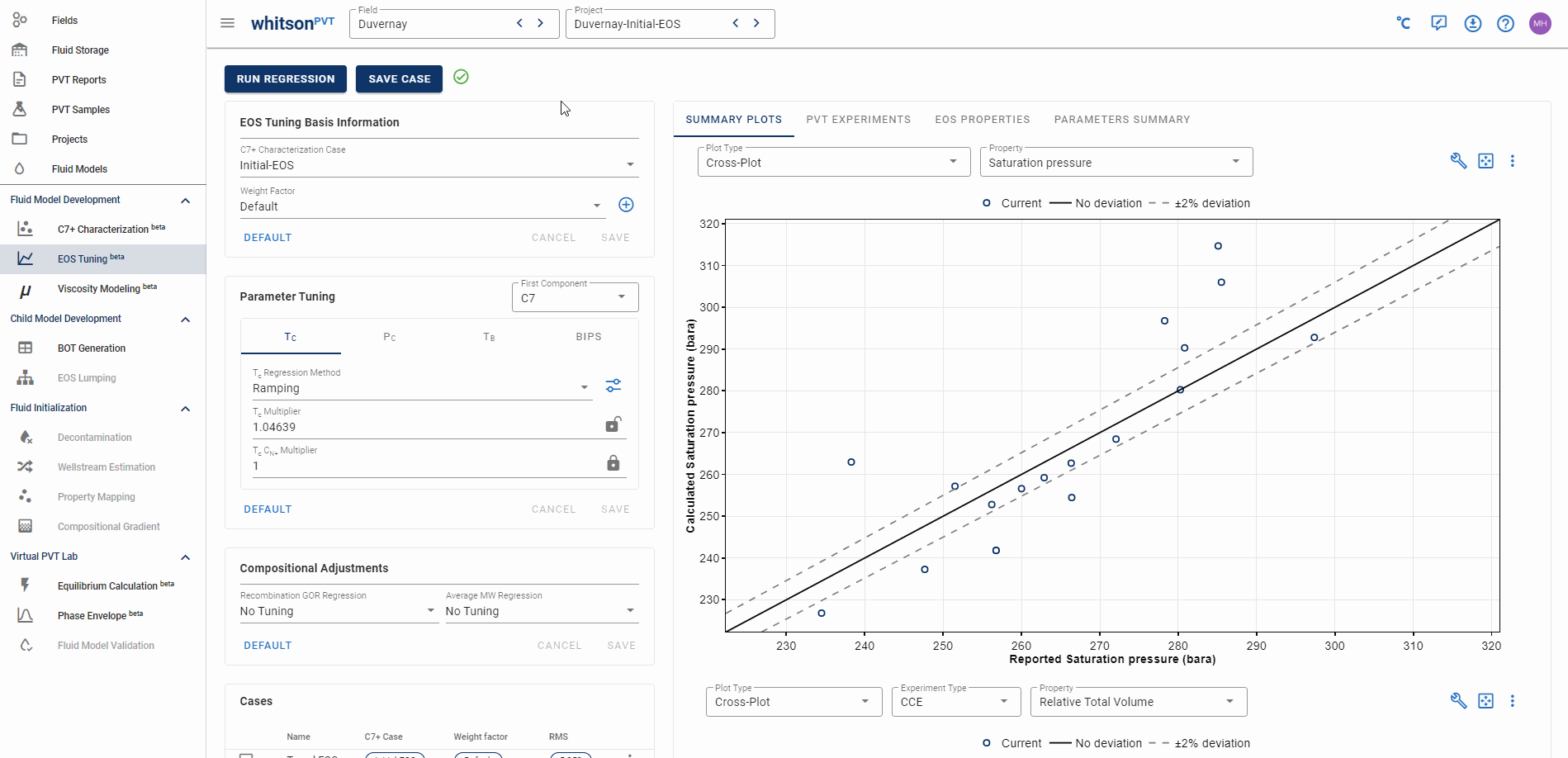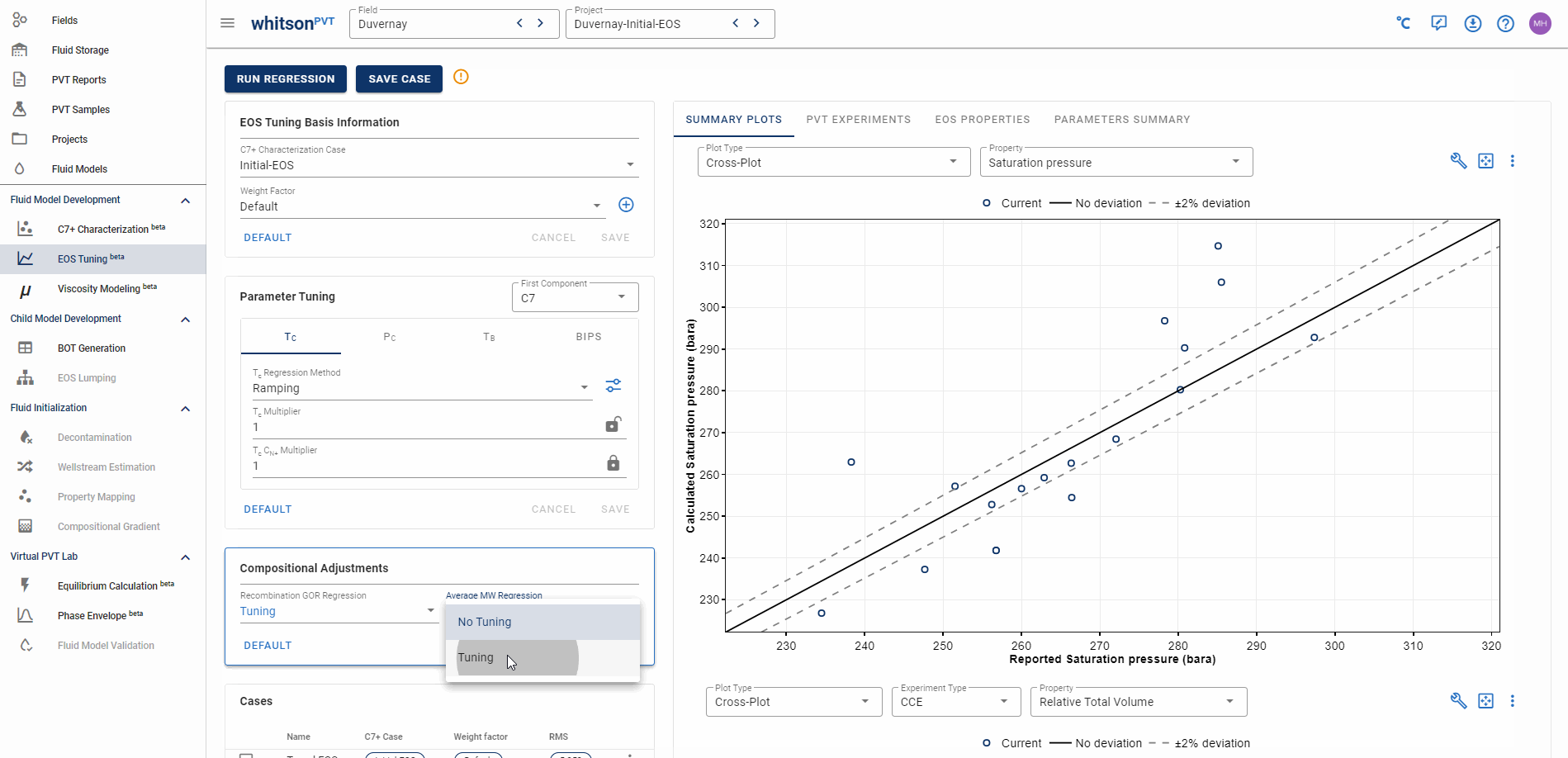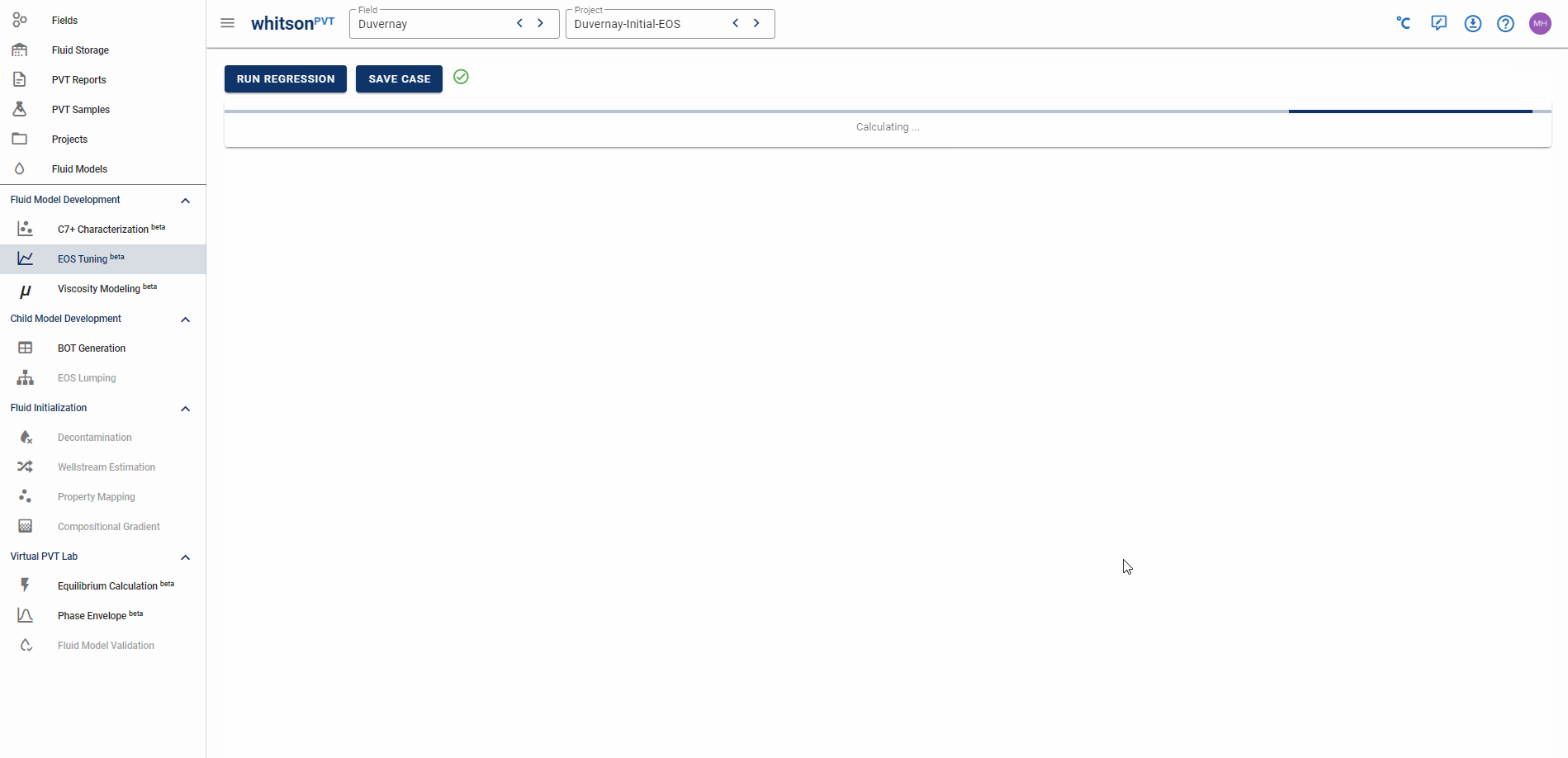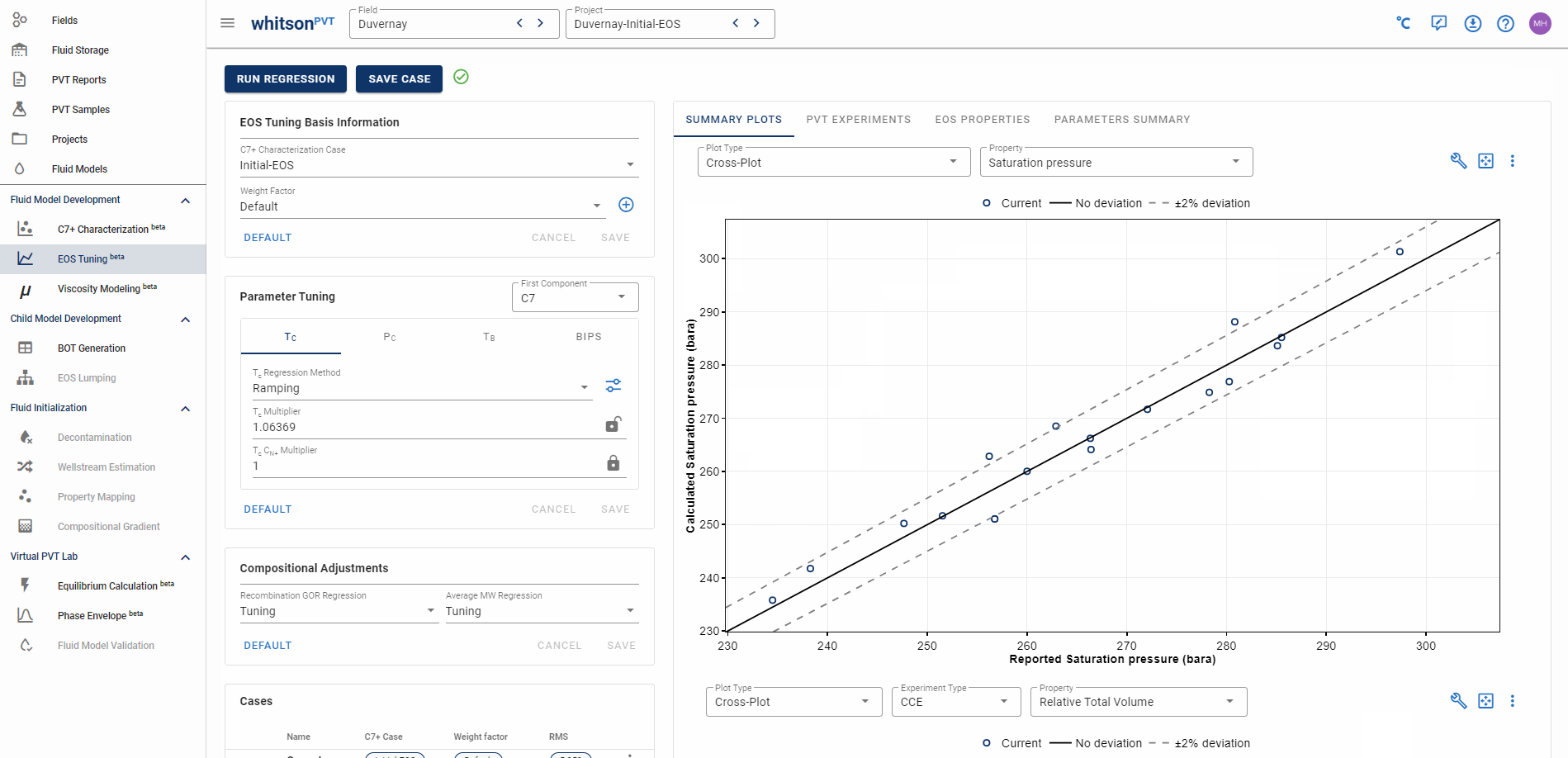
The top summary plots are there as a suggestion to help you check you model tuning fit! We recommend going from top to bottom when comparing tuning cases!
Some other plots you might want to check:
-
Multi-sample DLE data - In particular the oil formation volume factor and removed gas.
-
Multi-sample MSS data - Same data as the DLE and the oil density.
-
Induvidual sample CVD liquid dropout data.
-
Induvidual sample CCE liquid dropout data.
The EOS tuning predictions are divided into four tabs on the right-hand side of the feature. Each tab has a specific use and are explained below.
The Summary Plots is composed of two plots that both show multi-sample data predictions. There are three ways to display multi-sample data in whitsonPVT. The first is a cross-plot that compares the lab reported data on the x-axis and the EOS predicted data on the y-axis. The second type is a by sample deviation plot, that shows the deviation defined by Equation \eqref{eq:deviation_definition} for individual samples. The third type is a by property deviation plot that lets you identify at what reported values your deviation starts to increase or decrease!
The first summary plot has a set of whitson recommended properties that we recommend that you inspect and make a judgement about the quality of. The second (lower) summary plot is a by experiment summary plot for all samples that have data for that experiment type.
The second tab called PVT Experiments shows the experimental data and adjusted sample composition for induvidual samples. The top plot in this section lets you select a sample and its assosiated PVT experiment. Then you can change the x- and y- properties freely. The second plot in this section shows the mole composition of this sample with compositional adjustments and characterized component molecular weights converting from mass to mole amounts. The adjusted composition is in the detailed EOS component slate!
The third tab is called EOS Properties and just like in the C7+ Characterization feature, this section lets you see the tuning case EOS properties with a basis (x-axis) of molecular weight, normal boiling point, or component name. This lets you quickly check your tuned properties for thermodynamic consisitency, either by looking for non-monotonic behavior or if the data goes outside the pure compound properties (grey circles).
The forth and last tab called Parameters Summary lets you compare the EOS tuning parameters for your different cases! This is a quick way to see how your parameter tuning are affected for different cases.
What's Next?
Having developed one or more EOS Tuning cases, the next step is to develop a viscosity model! You can read about how to do this here.
Do you have any questions or comments? Feel free to reach out to our support email: support.pvt@whitson.com.